import numpy as np
import zarr
from matplotlib import pyplot as plt
from moraine.utils_ import is_cuda_available
if is_cuda_available():
import cupy as cp
from cupyx.scipy.ndimage import uniform_filter
import moraine as mrAdaptive Multilook
In this tutorial, we demostrate how to use Moraine package to identify spatially homogeneous pixels, extimate the coherence matrix and compare the original interferogram, multilook intergerogram and the adaptive multilook interferogram.
Load rslc stack
if is_cuda_available():
print(cp.cuda.Device(1).use())
rslc = cp.asarray(zarr.open('../../data/rslc.zarr',mode='r')[:])
print(rslc.shape)<CUDA Device 1>
(2500, 1834, 17)Apply ks test
if is_cuda_available():
rmli = cp.abs(rslc)**2
az_half_win = 5
r_half_win = 5
az_win = 2*az_half_win+1
r_win = 2*r_half_win+1
p = mr.ks_test(rmli,az_half_win=az_half_win,r_half_win=r_half_win)CPU times: user 216 ms, sys: 65.8 ms, total: 282 ms
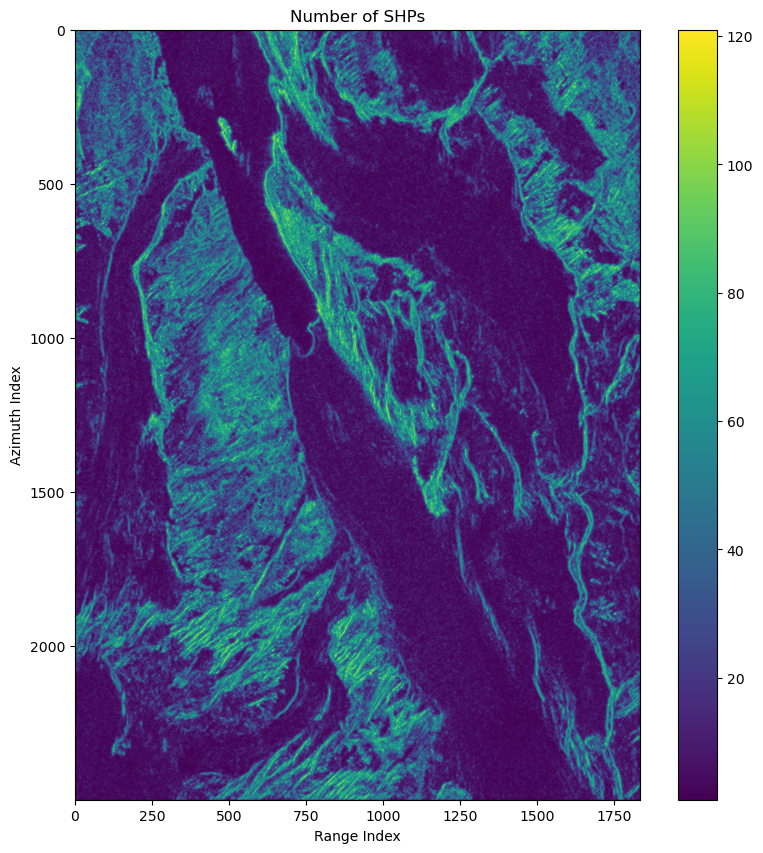
Wall time: 2.56 sSelect SHPs
if is_cuda_available():
is_shp = (p < 0.05) & (p >= 0.0)if is_cuda_available():
shp_num = cp.count_nonzero(is_shp,axis=(-2,-1))
shp_num_np = cp.asnumpy(shp_num)Estimate coherence matrix
if is_cuda_available():
coh = mr.emperical_co(rslc,is_shp)[1]CPU times: user 127 ms, sys: 20.6 ms, total: 148 ms
Wall time: 151 msCompare
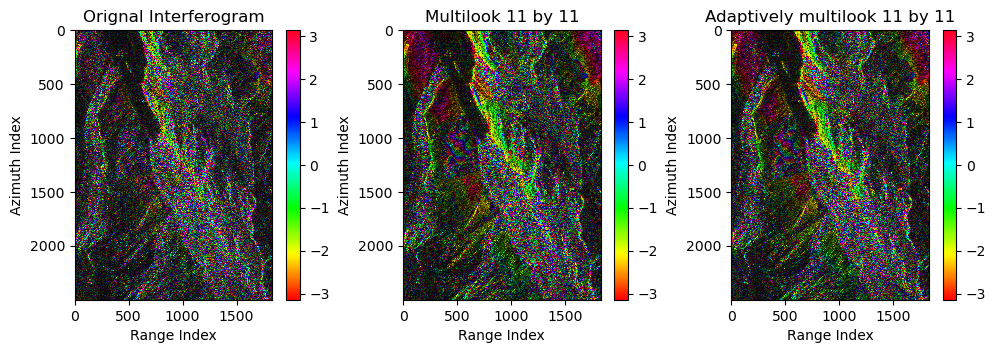
Here we compare 1-look interferogram, multilook interferogram and adaptive multilook interferogram
ref_image = 15
sec_image = 161 look interferogram:
if is_cuda_available():
diff = rslc[:,:,ref_image]*rslc[:,:,sec_image].conj()Multilook interferogram:
if is_cuda_available():
ml_diff = uniform_filter(diff,size=(az_win,r_win))Adaptive multilook interferogram:
if is_cuda_available():
ad_ml_diff = coh[:,:,ref_image,sec_image]The plot background:
if is_cuda_available():
plot_bg = rmli[:,:,0]
plot_bg = cp.asnumpy(plot_bg)
plot_bg = np.nan_to_num(plot_bg)
alpha = mr.bg_alpha(plot_bg)Plot:
if is_cuda_available():
fig,axes = plt.subplots(1,3,figsize=(24/2,7/2))
xlabel = 'Range Index'
ylabel = 'Azimuth Index'
pcm0 = axes[0].imshow(cp.asnumpy(cp.angle(diff)),alpha=alpha,interpolation='nearest',cmap='hsv')
pcm1 = axes[1].imshow(cp.asnumpy(cp.angle(ml_diff)),alpha=alpha,interpolation='nearest',cmap='hsv')
pcm2 = axes[2].imshow(cp.asnumpy(cp.angle(ad_ml_diff)),alpha=alpha,interpolation='nearest',cmap='hsv')
for ax in axes:
ax.set(facecolor = "black")
axes[0].set(title='Orignal Interferogram',xlabel=xlabel,ylabel=ylabel)
axes[1].set(title=f'Multilook {az_win} by {r_win}',xlabel=xlabel,ylabel=ylabel)
axes[2].set(title=f'Adaptively multilook {az_win} by {r_win}',xlabel=xlabel,ylabel=ylabel)
fig.colorbar(pcm0,ax=axes[0])
fig.colorbar(pcm1,ax=axes[1])
fig.colorbar(pcm1,ax=axes[2])
fig.show()Conclusion
- Adaptive multilooking based on SHPs selection performs better than non-adaptive one;
ks_testandemperical_coimplemented in Moraine package are fast.